LSU DeepDrug Team Uses Artificial Intelligence to Discover New Treatments for Coronavirus
Goal: New COVID-19 treatments within days or weeks.
The LSU DeepDrug team, a current semifinalist for the IBM Watson AI X-PRIZE, is using artificial intelligence, or AI, to discover new drugs. Although their work so far has focused mainly on antibiotics and antimicrobials, they are now working as quickly as they can to find new antivirals effective against coronavirus. The team is in the process of retraining their AI by feeding it large datasets of information and asking it questions—some with known answers—to see how well it is learning. Within days, they hope to have their AI begin suggesting new drugs, or drug combinations, to help save lives.

The LSU DeepDrug team brings together researchers Michal Brylinski, recent Rainmaker Award winner and associate professor in the Department of Biological Sciences with a joint appointment in the Center for Computation & Technology; Supratik Mukhopadhyay, associate professor in the Department of Computer Science, and Adam Bess, computer science graduate student
Some of the drugs the AI will suggest would need to go through rigorous testing to
show they’re safe and effective, but some would not. There are about 90 antiviral
drugs on the market that have already been approved by the FDA. It is possible that
DeepDrug’s AI will propose using one or a combination of them to treat COVID-19. This
could altogether eliminate the need for clinical trials and significantly push up
the timeline for getting drugs to patients. Most of the FDA-approved antivirals available
have so far been used to treat HIV, hepatitis B and C, and herpes, but could prove
effective against coronavirus as well.
To train their AI, DeepDrug is using a dataset of 2,700 antiviral peptides and associated
targets. The reason for this is the limited number of FDA-approved antivirals that
target influenza and positive-strand RNA-type viruses, which coronaviruses are. Available
drugs simply don’t provide enough data to render the artificial intelligence “smart.”
“We cannot eliminate the virus from the body or prevent it from infecting more people. What we can do is lower the threat and lower the mortality rate—especially for people with severe conditions who instead could have mild conditions. They’ll still get infected, but they’ll survive. More people could survive the pandemic.”—Michal Brylinski
One component of DeepDrug’s software is called eDrugRes, which the team has been using
to predict drug resistance in bacteria. But the way eDrugRes works, by analyzing the
protein-protein interactions in cells, can be applied to viruses as well.
“Earlier, we looked at the protein-protein interactions across many different species, actually
18,000 different bacterial strains,” said Adam Bess, an LSU computer science graduate
student who is working with lead researchers Supratik Mukhopadhyay, associate professor
in the Department of Computer Science, and Michal Brylinski, recent Rainmaker Award winner and associate professor in the Department of Biological Sciences with a joint appointment
in the Center for Computation & Technology. “Now, we’re only analyzing the human side;
the different mechanisms through which viruses move and propagate in the body. Because
of the lack of data, we have to use a slightly different methodology.”
The team, with guidance from LSU immunotoxicologist Stephania Cormier, is analyzing
45 known mechanisms, including how a virus forms a bond with a host cell; membrane
fusion, the process through which a virus enters a cell; how a virus unpacks its contents;
how it replicates itself; and how new copies of the virus then go infect other host
cells.
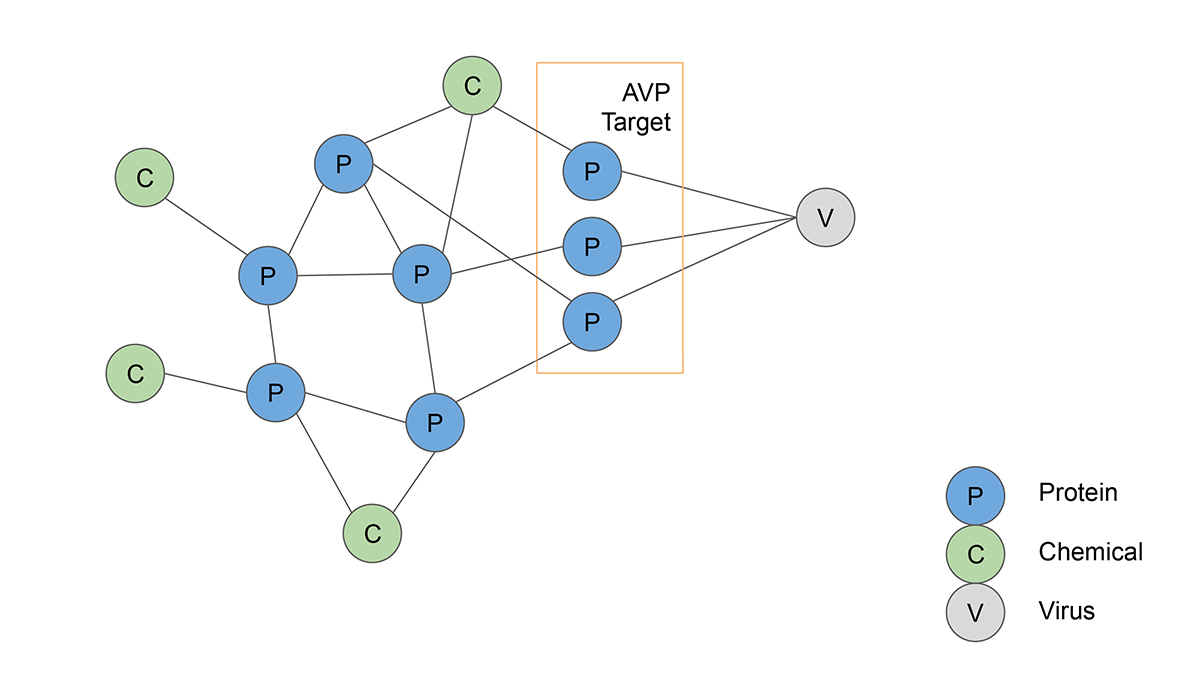
Artificial intelligence helps map known antiviral peptides (AVPs) to different cell mechanisms by analyzing protein-protein interactions. The AVPs are then ranked by how effectively they can slow the propagation of coronavirus in the human body.
“Essentially, we’re looking at all of the different aspects of how a virus spreads
throughout the body,” Mukhopadhyay continued. “And then we link these antiviral peptides
with each of these mechanisms, associate them with specific protein pathways, and
score them depending on how effective they are at inhibiting a particular mechanism.
At the end, we should know which are the three or four drugs best suited to slow or
prevent a virus from propagating.”
“The mapping is a critical component,” Brylinski added. “If we cannot map the peptides
properly, nothing is going to work. But if we can, we’ll establish very good connections
between viral activity and specific drugs in one unifying framework. It’s challenging
but exciting work.”
If the team is successful, their AI could suggest antiviral drugs to reduce the impact
of COVID-19 within mere days. The team is careful to point out, however, that their
end result won’t be neither a vaccine nor a complete cure.
“We cannot eliminate the virus from the body or prevent it from infecting more people,”
Brylinski clarified. “What we can do is lower the threat and lower the mortality rate—especially
for people with severe conditions who instead could have mild conditions. They’ll
still get infected, but they’ll survive. More people could survive the pandemic.”
A few drugs, such as chloroquine (and its derivative hydroxychloroquine, both used
to prevent and treat malaria) along with azithromycin and remdesivir, have been approved
by the FDA for treating SARS-CoV-2 (betacoronavirus) infections. Most of these treatments
were discovered through trial and error in different parts of the world. What DeepDrug
proposes is a principled approach to drug discovery and drug repurposing (meaning,
established drugs are used to treat new or different conditions) based on datasets
that would be too large—or at least quite slow—to process without the use of artificial
intelligence. Also, by helping to predict the success/failure rate of a drug, the
AI could prevent researchers from wasting a lot of time exploring what are likely
to be dead ends. DeepDrug’s AI would save time exploring the currently FDA-approved
antivirals to see if they could treat COVID-19 as well—along with many possible combinations
with other antivirals and drugs.
Elsa Hahne
LSU Office of Research & Economic Development
225-578-4774
ehahne@lsu.edu